Gladstone NOW: The Campaign Join Us on the Journey✕

Gladstone researcher Alex Pico helped develop a new way to measure the difference one discovery can make on science around the world.
Five years ago, a business-minded potential donor asked Sandy Williams, who was president of Gladstone at the time, how someone might measure the return on investment a donor could expect from a contribution to discovery science.
Williams intuitively knew that the kinds of biomedical breakthroughs that donors hope for, such as drugs or devices, arise from the work of many people over many years at multiple institutions. But could he quantify it?
Cure Networks
After mulling over the donor’s question about return on investment, Williams approached the heads of the bioinformatics core at Gladstone, including Pico, PhD, associate director of bioinformatics. Pico recognized that scientific research results in incremental gains in knowledge that eventually lead to cures, devices, or other noteworthy returns on investment. And, to Pico, the idea of tracing how this happens over time looked like a network problem.
Pico was accustomed to studying biological networks—interactions among the components of cells, such as genes and proteins. But the tools he used were equally applicable to other types of networks, such as social networks or water distribution networks. He reasoned that these tools could also apply to citation networks—papers citing other papers that cite other papers. Pico and his collaborators hypothesized that such networks might reveal the extent to which a broad base of collaborative research actually lies behind important biomedical success stories.
To test their hypothesis, they asked: Just how many researchers, institutions, and collaborations contribute to a single cure? Using data mining and a network visualization software called Cytoscape, they extracted from the repository of published medical literature (PubMed), the citation network behind the FDA approval of two new drugs.
The papers cited in the FDA submissions for a drug are apparent to anyone who reads those submissions. “But, you can’t go to PubMed and say, ‘Hey, tell me about all the papers that are one or two steps away from this set of clinical trials,” Pico said. “No one has made that database available.”
To address that gap, Pico and his team identified, for each drug, two generations of cited papers backward in time. For one drug, the network they extracted included 7067 researchers from more than 5000 institutions, working over the course of 104 years. Results for the second drug were similar: the citation network spanned 59 years and included 2857 different scientists with 2516 different institutional and departmental affiliations. These findings suggest that successful breakthroughs rely on a broad base of research rather than just a few high-impact papers.
“I was surprised by the scale of it in both time and numbers,” Pico said. “My intuition was that it would be a more selective club.”
He wasn’t the only one who was impressed. After the work was published in the journal Cell, National Institutes of Health (NIH) Director Francis Collins cited the findings during his 2016 testimony before a congressional committee, urging support of basic science research.
In addition, George Chacko, chief scientific officer at Net ESolutions (NETE), found the work was one of the most interesting research articles he had read in 10 years. “From the science history perspective, it was unbelievable. You can go back and see how interactive science is. How all this work culminated in something at the top of the pyramid—for example, a drug approved for human use.”
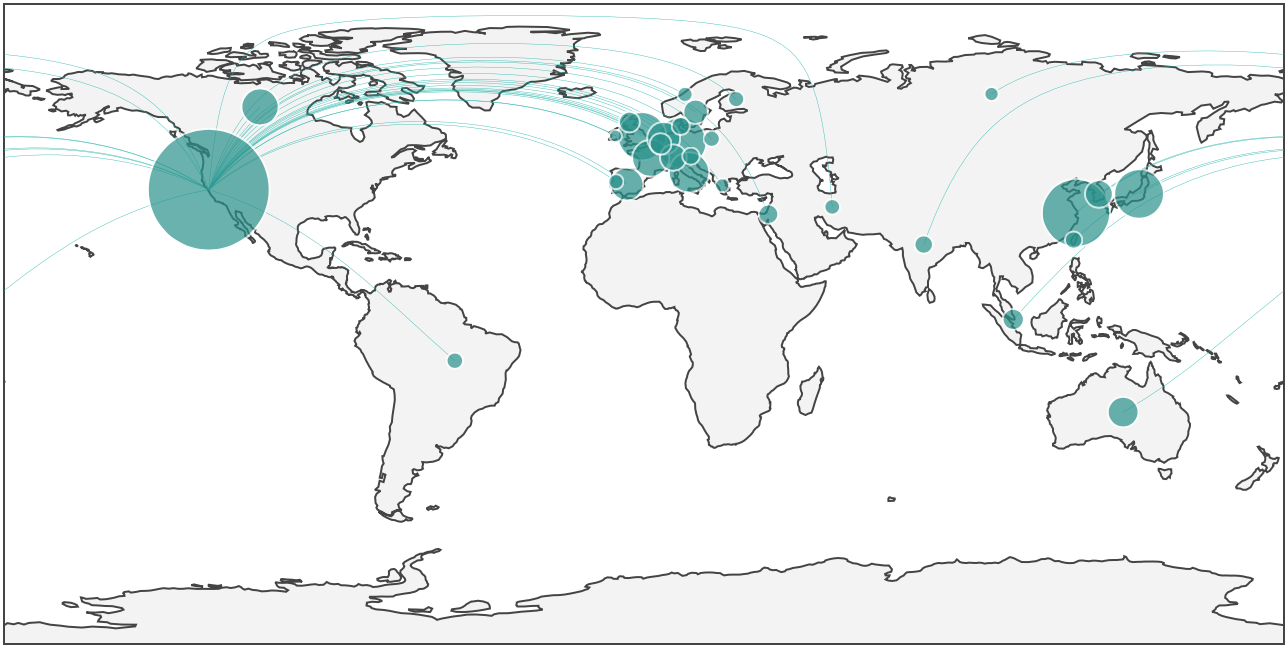
Gladstone’s research papers have been cited by authors all around the world. The size of each dot relates to the number of publications in that country that have cited a core set of 220 Gladstone stem cell papers.
To Chacko, the work also suggested an inspirational path forward for NETE, which provides digital design, development, and management services solutions to US federal agencies, including the NIH. For example, citation networks might be used to evaluate any organization’s or individual’s research portfolio. So, he fired off an email to Pico, and a collaboration between Gladstone and NETE was born.
The team’s initial project retrieved citations pertaining to five FDA-approved cancer therapies. They fleshed out their data set to include clinical research papers, FDA regulatory submissions, patent documents, and post-drug approval literature reviews for the five drugs, and then searched backward through two generations of cited papers. In the end, they extracted a citation network consisting of more than 100,000 papers authored by over 235,000 people.
Only 14 publications were cited in all five of the drugs’ citation networks, and all of them related to basic science endeavors, an indication that much value lies in basic research that is not specifically geared toward a particular end product. The researchers also found more than 19,000 NIH grants tied to the 100,000+ publications in the citation network. Over 100 of those grants were found in all five networks, reinforcing the impression that successful drug development depends on investment in extensive collaboration and broad community engagement.
Although this work was focused on the citation networks behind cures, the team recognized that the same technique could be used to derive a citation network for a medical device, a Nobel prize, or any other scientific success story. The approach might even be flipped into a forward-looking citation network that can quantify the impact of research by an individual, department, or university on a particular discipline or across a range of disciplines.
A Small Business Collaboration
Based on its initial collaboration with Gladstone, NETE saw a potential business opportunity. Perhaps a citation network–based knowledge platform could address the need of funding organizations to justify the value of their research programs to stakeholders.
With this in mind, Chacko and NETE, with Pico as a consultant, applied for and received a National Institute on Drug Abuse Small Business Innovation Research (SBIR) Program award to build such a platform, which they named ERNIE (Enhanced Research Network Informatics Environment). ERNIE’s core is a database of diverse research metadata from public and commercial sources, as well as modular tools for extracting data and generating networks for analysis.
ERNIE is a much-expanded version of the original Gladstone work, with bigger and better data sets and reliable, effective computer code, Chacko said. However, the underlying theme of defining a core set of publications and their network of citations persists. “It’s our database and code now and works slightly differently, but the concept is the same,” he said.
Gladstone’s Stem Cell Research: The Alpha-User Test
Before taking ERNIE on the road, Chacko and Pico gave it one more test drive at Gladstone to make sure ERNIE’s data set, workflows, and program features were ready for prime time. In a 3-month alpha-user test, they set out to evaluate the impact of the Gladstone stem cell program. “Gladstone already has a tremendous reputation for stem cell work,” Chacko said, “but it’s nice to have evidence, even to support the things you’re sure of.”
Unlike the cure networks, where it made sense to extract citations from the top down (going backward in time from the cure), “our idea for the alpha-user test was to combine both the top-down and bottom-up approaches,” Pico said.
They started by searching for all Gladstone papers that used the terms “stem cell” and “iPS cell” (induced pluripotent stem cell) from 2002 to 2018. For the 220 papers they found, they then built citation networks that extended two citation generations both backward and forward in time from each paper.
The results confirmed that Gladstone has both a national and global footprint in iPS cell research. The 220 Gladstone stem cell papers have been cited more than 16,000 times. And 14 of the Gladstone papers have accumulated more than 200 citations each in the 10 years since publication—a number that ranks them in the top .06 percent of most cited papers, according to Chacko.
This work offered a few hints of what ERNIE might be able to do in the future. For example, an organization could dig deeply into how its research has been used by others, and might even use that information to inform its strategic planning. Indeed, NETE is now moving forward with ERNIE, combining Gladstone’s citation network approach with other techniques built with other collaborators.
Pico’s citation networks have come a long way since meeting their initial goal of better understanding the impact of basic science. At the start, he saw it as a fairly focused side project. “We weren’t thinking at all how this might blossom into business opportunities and future grants,” Pico said. But that’s kind of the nature of basic science research. It can take off in many directions, some of them unforeseen. “This sort of serendipity is familiar to those of us working in research fields,” he said, “but it’s never taken for granted.”
Support Discovery Science
Your gift to Gladstone will allow our researchers to pursue high-quality science, focus on disease, and train the next generation of scientific thought leaders.
New Tool Reveals the Secrets of HIV-Infected Cells
New Tool Reveals the Secrets of HIV-Infected Cells
Developed by Gladstone scientists, HIV-seq could uncover new opportunities for treating HIV.
News Release Research (Publication) HIV/AIDS Infectious Disease Roan LabVitamin B3 Therapy Offers Hope for Fatal Childhood Disease
Vitamin B3 Therapy Offers Hope for Fatal Childhood Disease
A new framework that matches vitamins with genetic diseases helped uncover that high-dose vitamin B3 can dramatically extend survival in mice with NAXD deficiency.
News Release Research (Publication) Rare Diseases Jain LabRed Blood Cells Soak Up Sugar at High Altitude, Protecting Against Diabetes
Red Blood Cells Soak Up Sugar at High Altitude, Protecting Against Diabetes
New study shows red blood cells act as hidden glucose sponges in low-oxygen conditions, explaining why people living at high altitude have lower diabetes rates and pointing toward new treatments.
News Release Research (Publication) Diabetes Jain Lab