The Gladstone Flow Cytometry Core enables scientists to analyze or isolate cells based on their phenotypic or functional properties, and can be used in conjunction with other core technologies to form a robust and effective pipeline for cellular analysis. Analysis of up to 18 and sorting of up to 15 fluorescent parameters is available from a wide variety of samples and cell types. Our staff is experienced in experimental and antibody panel design, practical operation of multiple flow cytometric platforms, and basic and advanced data analysis.
Discover how our core can help you with your flow cytometry needs.
Services Provided
Cell Sorting
Cell sorting for up to 18 fluorescent parameters with the following specifications:
- 5 available lasers: UV (355nm), Violet (405nm), Blue (488nm), Yellow-Green (561nm), Red (640nm).
- Functional marker sorting and analysis (e.g., phospho-flow, cell cycle, and mitochondrial assays).
- Single-cell sorting into 96-well plates for cloning or further sequencing downstream analysis.
Sorting into the following receptacles:
- 1.5ml Eppendorf tubes
- 5ml Falcon tubes
- 15ml Falcon tubes
- Any well plate (12, 24, 48, 96 and 384)
- Terasaki plate
- Standard/frosted end slide
Aseptic sorting is available.
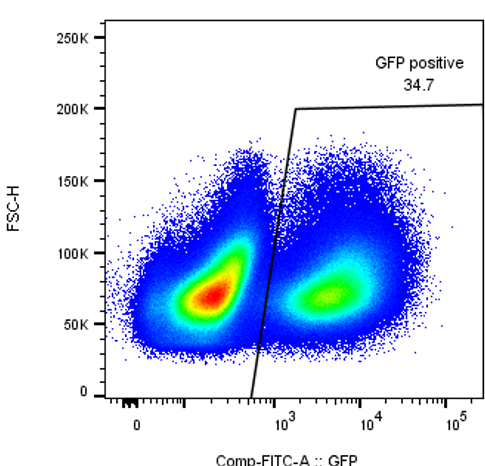
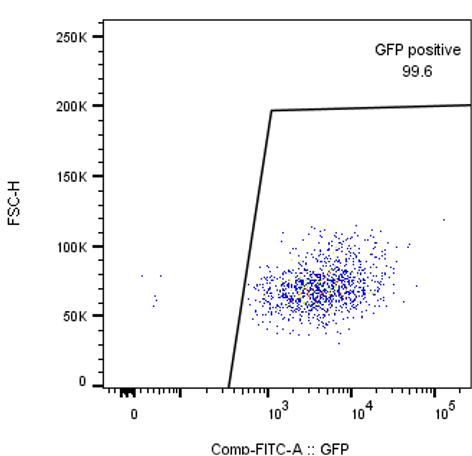
Purification of GFP positive cells using the FACSAria Fusion.
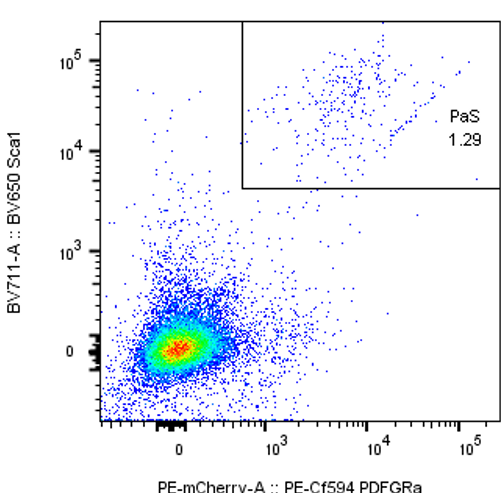
Detection of mouse mesenchymal PaS cells.
Cell Analyzers
- Analysis of up to 40 fluorescences in one tube.
- Analysis of extra, intracellular and functional markers as well as fluorescent proteins.
- High-throughput analysis from 96- and 384-well plates (up to 18 fluorescent parameters) for all analyzers.
Imaging Cytometry
- Imaging Cytometry for up to 6 fluorescences, or 4 fluorescences including brightfield and side scatter.
- 20X, 40X and 60X magnification available.
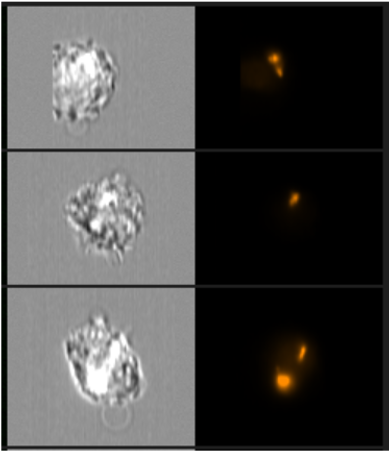
Detection of mCherry positive bacteria in cells using the Imagestream.
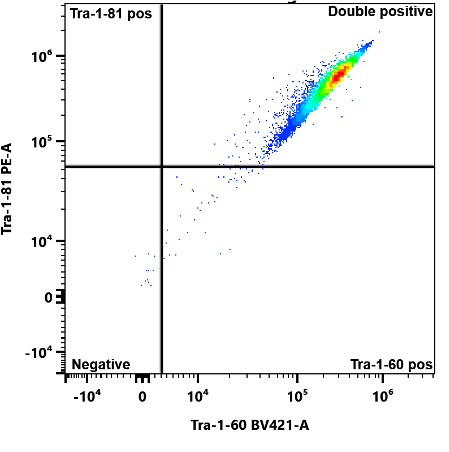
Detection of pluripotency markers Tra-1-60 and Tra-1-81 in human induced pluripotent stem cells.
hiPSC Pluripotency Testing
Test whether the human induced pluripotent stem cells (hiPS cells) that you have received or grown express a panel of pluripotent markers and define their pluripotency status. Markers include Tra-1-60, Tra-1-81, SSEA-3, SSEA-4, CD30, CDH3, Nanog, Oct 3_4 and Sox-2, which you can choose from to run in your panel. Turnaround time is approximately 1 day and you’ll receive all raw data files at the conclusion of the experiment.
Additional Services
Gladstone’s Flow Cytometry Core also offers sample processing, including staining and running of experimental samples. Data analysis services are also available.
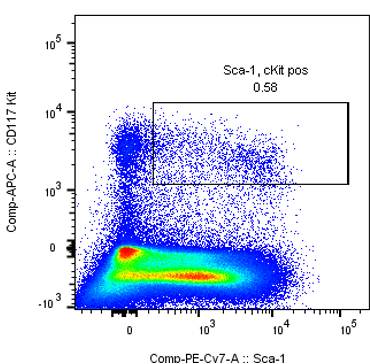
Detection of Sca-1/c-Kit positive markers in mouse hematopoietic stem cells.
Fees and Scheduling
For more information on our fees, contact the core.
Publications
A molecular mechanism for probabilistic bet hedging and its role in viral latency. Sonali Chaturvedi, Jonathan Klein, Noam Vardi, Cynthia Bolovan-Fritts, Marie Wolf, Kelvin Du, Luwanika Mlera, Meredith Calvert, Nathaniel J. Moorman, Felicia Goodrum, Bo Huang, Leor S. Weinberger.
HIV efficiently infects T cells from the endometrium and remodels them to promote systemic viral spread. Tongcui Ma, Xiaoyu Luo, Ashley F George, Gourab Mukherjee, Nandini Sen, Trimble L Spitzer, Linda C Giudice, Warner C Greene, Nadia R Roan.
SIRT1 is downregulated by autophagy in senescence and ageing. Caiyue Xu, Lu Wang, Parinaz Fozouni, Gry Evjen, Vemika Chandra, Jing Jiang, Congcong Lu, Michael Nicastri, Corey Bretz, Jeffrey D. Winkler, Ravi Amaravadi, Benjamin A. Garcia, Peter D. Adams, Melanie Ott, Wei Tong, Terje Johansen, Zhixun Dou, Shelley L. Berger.
Remote Training of SRL Users and Staff in a Global Pandemic. Kathleen Daniels, Alexis Conway, Rui Gardner, Lola Martinez, Kylie M. Price, Sarah Schneider, Rachael Sheridan, Jane Srivastava, Sherry Thornton.
Shared Resource Laboratory Operations: Changes Made During Initial Global COVID‐19 Lockdown of 2020. Jessica B. Back, Cora H. Chadick, Juan J. Garcia Vallejo, Eva Orlowski‐Oliver, Radhika Patel, Caroline E. Roe, Jane Srivastava, Rachael V. Walker.
Transcription Factor Overexpression Drives Reliable Differentiation of Retinal Pigment Epithelium from Human Induced Pluripotent Stem Cells. Tessa E. Dewell, Ketrin Gjoni, Angela Z. Liu, Ashley R.G. Libby, Anthony T. Moore, Po-Lin So, Bruce R. Conklin.
Shared Mechanisms Govern HIV Transcriptional Suppression in Circulating CD103+ and Gut CD4+ T Cells. Steven A. Yukl, Shahzada Khan, Tsui-Hua Chen, Martin Trapecar, Frank Wu, Guorui Xie, Sushama Telwatte, Daniel Fulop, Alexander R. Pico, Gregory M. Laird, Kristen D. Ritter, Norman G. Jones, Chuanyi M. Lu, Robert F. Siliciano, Nadia R. Roan, Jeffrey M. Milush, Ma Somsouk, Steven G. Deeks, Peter W. Hunt, Shomyseh Sanjabi.
FOXO1 promotes HIV Latency by suppressing ER stress in T cells. Albert Vallejo-Gracia, Irene P. Chen, Rosalba Perrone, Emilie Besnard, Daniela Boehm, Emilie Battivelli, Tugsan Tezil, Karsten Krey, Kyle A. Raymond, Philip A. Hull, Marius Walter, Ireneusz Habrylo, Andrew Cruz, Steven Deeks, Satish Pillai, Eric Verdin, Melanie Ott.
Silencing of E-cadherin in induced human pluripotent stem cells promotes extraembryonic fates accompanying multilineage differentiation. Ashley RG Libby, Ivana Vasic, David A Joy, Martina Z Krakora, Fredrico N Mendoza-Camacho, Bruce R Conklin, Todd C McDevitt.
In Vivo Chimeric Alzheimer’s Disease Modeling of Apolipoprotein E4 Toxicity in Human Neurons. Ramsey Najm, Kelly A.Zalocusky, Misha Zilberter, Seo Yeon Yoon, Yanxia Hao, Nicole Koutsodendris, MaxineNelson, Antara Rao, Alice Taubes, Emily A. Jones, Yadong Huang.
Training
How Do I Get Trained to Use the Equipment in the Gladstone Flow Cytometry Core?
Contact the flow core staff to start the practical training process.
To get started, you’ll need to:
- Register for an iLab account.
- Read, sign, and return the signature page of the general regulations document to the core by email.
- Complete Gladstone’s Flow Core Intro Questionnaire.
You can also refer to additional resources to learn more about flow cytometry.
Support your research needs with our cutting-edge equipment and expertise.

NanoFCM Analyzer
The NanoFCM Analyzer can be used for the multiparameter characterization, sizing and concentration of natural and synthetic nanoparticles (7-1000 nm) at the single-particle level, including detection of extracellular vesicles, mitochondria, bacteria, viruses, nanomedicine, nanomaterial and other nanoparticles.
Specifications:
- Laser (blue, green, red)
- One side-scatter channel
- Two fluorescence channels
- Up to 12,000 particles/min
- Lower detection limit 7 nm (gold nanoparticles) to 40 nm (exosomes, viruses)
- High detection limit (1 micron)
Practical and theoretical training is available for the Nano FCM, as well as experimental and data analysis support.

Fortessa X-20
The Fortessa X-20 cell analyzer delivers high-performance, multicolor analysis and is configured with five lasers to detect up to 20 parameters (18 fluorescent and 2 scatter) simultaneously. An integrated High Throughput Sampler (HTS) provides rapid, fully automated sample acquisition from 96- and 384-well microtiter plates.
Practical and theoretical training is available for the Fortessa X-20, as well as experimental and data analysis support.
| LASER | CHANNEL | BANDPASS FILTER | LONGPASS MIRROR |
|---|---|---|---|
| UV (355 nm) | BUV737 | 740/35 | 690LP |
| BUV496 | 515/30 | 450LP | |
| BUV396 | 379/28 | ||
| Violet (405 nm) | BV786 | 780/60 | 750LP |
| BV711 | 710/50 | 690LP | |
| BV650 | 670/30 | 635LP | |
| BV605 | 610/20 | 600LP | |
| BV510/AMCYAN | 470/15 | 450LP | |
| BV421/BFP/DAPI | 431/28 | 410LP | |
| Blue (488 nm) | Per-CP-Cy5.5 | 695/40 | 690LP |
| FITC, BB515 | 530/30 | 505LP | |
| SSC | 488/10 | ||
| FSC | |||
| Yellow/Green (561 nm) | PE-Cy7 | 780/60 | 750LP |
| PE-Cy5 | 670/30 | 650LP | |
| PE-CF594 | 610/20 | 600LP | |
| PE | 586/15 | 570LP | |
| Red (637 nm) | APC-CY7 | 780/60 | 750LP |
| AF-700 | 710/50 | 690LP | |
| APC | 670/30 | 650LP |

Cytek Aurora
The Cytek Aurora is a spectral cytometry analyzer configured with four lasers (violet, blue, yellow/green, and red) to detect up to 48 parameters simultaneously. Other features include autofluorescence extraction, ability to determine fluorescences in one tube that are unable to be distinguished by regular flow cytometry, and intuitive software with transferable experimental workspaces.
Practical and theoretical training is available for the Aurora, as well as experimental and data analysis support.
Aurora configuration:
- 16 violet parameters
- 14 blue parameters
- 10 yellow/green parameters
- 8 red parameters

BD FACSAria Fusion Cell Sorter
Two BD FACSAria Fusions cell sorters are configured to sort up to 20 parameters (18 fluorescence and 2 scatter). A choice of nozzles allows users sort a wide range of particle sizes. Nozzles are available in four sizes: 70, 85, 100, and 130 microns. Bulk, single cell and index sorting are available.
Practical and theoretical training is available for the Fusion, as well as experimental and data analysis support.
| TUBES | PLATES | SLIDES |
|---|---|---|
| 5 ml Falcon | 6 well | Standard |
| 15 ml Falcon | 12 well | Frosted End |
| Eppendorf | 24 well | |
| 48 well | ||
| 96 well | ||
| 384 well | ||
| Terasaki |
| LASER | CHANNEL | BANDPASS FILTER | LONGPASS MIRROR |
|---|---|---|---|
| UV (355 nm) | BUV737 | 730/45 | 690LP |
| BUV496 | 515/30 | 450LP | |
| BUV396 | 379/28 | ||
| Violet (402 nm) | BV786 | 780/60 | 750LP |
| BV711 | 670/30 | 690LP | |
| BV650 | 670/30 | 635LP | |
| BV605 | 610/20 | 600LP | |
| BV480 | 470/14 | 450LP | |
| BV421 | 431/28 | 410LP | |
| Blue (488 nm) | Per-CP-Cy5.5 | 710/50 | 690LP |
| FITC | 530/30 | 502LP | |
| SSC | 488/10 | ||
| FSC | |||
| Yellow/Green (561 nm) | PE-Cy7 | 780/60 | 750LP |
| PE-Cy5 | 670/30 | 650LP | |
| PE-CF594 | 610/20 | 600LP | |
| PE | 586/15 | 570LP | |
| Red (637 nm) | APC-CY7 | 780/60 | 750LP |
| AF-700 | 710/50 | 690LP | |
| APC | 670/30 | 650LP |

Attune NxT Flow Cytometer
The Attune NxT Flow Cytometer is a compact, benchtop cell analyzer that has been configured with four lasers to run and analyze multicolor panels of up to 16 colors (14 fluorescence and 2 scatter). An integrated autosampler provides rapid, fully automated sample acquisition from 96- and 384-well microtiter plates.
Practical and theoretical training is available for the Attune, as well as experimental and data analysis support.

Amnis Imagestream X Mark II
The Amnis Imagestream X Mark II is an imaging cytometer that combines the speed, sensitivity, and phenotyping abilities of flow cytometry with the detailed imagery and functional insights of microscopy. The Imagestream comes equipped with 20, 40 and 60X magnification, as well as the ability to detect up to 6 fluorescent channels (or 4 plus brightfield and side scatter) using 4 lasers (405, 488, 561 and 635nm). Cells can be run at up to 5,000 events per second.
Applications for the Imagestream include:
- Co-localization
- Internalization
- Cell cycle and mitosis
- Cell signaling and interaction
- Shape change and chemotaxis
- Nuclear translocation
Practical and theoretical training is available, as well as experimental and data analysis support.
Support Discovery Science
Your gift to Gladstone will allow our researchers to pursue high-quality science, focus on disease, and train the next generation of scientific thought leaders.
FAQs
How Do I Book a Session on a Flow Cytometer or Cell Sorter?
You’ll first need to register for an iLab account. Once that’s complete, you should read, sign, and return the signature page of the general regulations document to the core by email.
Contact core staff if you have not used the core before.
Which Flow Cytometers Are Capable of Running Plates?
The Fortessa X-20, Attune NxT, and Cytek Aurora, are capable of analyzing 96- and 384-well plates.
How Do I Get Trained to Use the Flow Cytometry Core?
If you'd like to be trained on an analyzer or sorter, complete Gladstone's Flow Core intro questionnaire to answer some questions about your experience, and email the core to schedule a date.
What Type of Samples Can Be Run on the Cytometer?
Any single-cell suspension where cells are between 500 nm and 20 μm can be run. Samples can be provided in 5-ml Falcon tubes, 15-ml Falcon tubes, Eppendorf tubes, multi-well plates, or slides.
Cells can be at a concentration of up to 5 million per mL in PBS or media, but serum should be limited to 2 percent serum, as more than this can affect the charge applied to the stream. You can also add HEPES (25 mM), EDTA (1 mM) or DNAse (10 U/ml) to avoid clumping. If you are concerned that your sample is too concentrated, bring in some media to dilute your sample. Filter your cells through a cell strainer or filter-top tube.
Can the Cytometers Be Used Independently, After Hours, or on Weekends?
After being trained, you must demonstrate proficiency in cytometer operation, maintenance, and basic troubleshooting before being allowed to use the equipment independently or outside of operating hours.
Does the Flow Core Archive Data?
The core saves a copy of the FCS files to an internal server.






