The Histology and Light Microscopy Core provides technical assistance, training, consultation, and collaboration for all aspects of experimental design, sample preparation, image processing, and data analysis. Our core is equipped with state-of-the-art technologies and expertise in histology, high-resolution histological imaging, confocal microscopy, light sheet microscopy, spinning disk microscopy, and optical projection tomography.
We strive to provide you with the knowledge and equipment you need to successfully perform your experiment. Our core also offers self service access for trained users.
Biology is a beautiful thing, and we want to work with you to make it that way!
Contact
Email@email for any information relating to POs, quotes, and administrative inquiries.
Email @email for any technical questions regarding projects.
Core Members
Rebecca Blandino, PhD
Senior Research Technologist
Rose Guerrero
Research Scientist
Alex Liu
Research Technologist I
Jane Srivastava
Scientific Resource Director
Services Provided
In addition to technical assistance and consultation on our services, we also provide training so that you can utilize the equipment on your own time.
Histology
- Tissue processing (paraffin/FFPE and frozen/cryo)
- Tissue embedding
- Tissue sectioning
- Tissue staining and whole slide scanning
- H&E
- IHC, IFA
- ISH, TUNEL
- Special staining (PAS, ORO, Congo Red, Masson's Trichrome)
- Spatial Biology: 10x Visium, RNAscope
Advanced Microscopy
On live and fixed samples
- Confocal/super-resolution
- Spinning disk
- Light-sheet, tissue clearing, volumetric imaging
- Automated whole slide scanning
- Widefield microscopy
Computer-Based Data Analysis
- Mammoth: Imaris Workstation (Room 327)
- Imaris Software
- Stitchy Software “Stitching Software”
- Imagescope, ImageJ/FIJI, OlyVia…
- Gladstone Institute of Cardiovascular Disease Workstation
- Gladstone Institute of Neurological Disease Workstation
- Stereology: StereoInvestor
Training, consulting, and collaborations
Self-services: Equipment Usage (after training)
If you have additional questions, check out our FAQs.
Fees and Scheduling
Scheduling
The Histology and Light Microscopy Core recharges researchers at an hourly rate for training and support with microscopes and sample preparation. Consult with our staff before preparing samples to ensure your reagents match the available equipment.
You are required to complete an initial training session before using any equipment in the core.
If you are part of UC San Francisco:
- Create an account on iLab
- Add your lab or PI’s PO number to your profile or account
- Request services on iLab
If you are part of a nonprofit institution or a private company:
- Contact the core at histology@gladstone.ucsf.edu
- Complete the lab service agreement and email it to the core
- Create an account on iLab
- Request services on iLab
Microscope Reservation
To start your session with an already existing reservation:
- To access your existing reservation, visit the equipment kiosk. Use your iLab credentials to log in.
- Once logged in, you will see a list of your pre-scheduled reservations under “My kiosk sessions.”
- Once you find your session, click the green “start” button. You will see the details of your reservation as well as a timer.
- You may log out while your session is in process. To log out, click the upper right-hand side menu and select "Log out." On the logout screen, you will see your list of Active sessions.
To start your session as a walk-in:
- Log in to the core kiosk using your iLab credentials.
- Select the instrument you would like to use from the menu.
- Once a calendar of availability appears, select “create session." Choose your desired duration, and click “create session” again.
- A new window will appear with the details for that reservation. You may be required to enter in your payment information and the equipment use type.
- Once you fill out all the required information, click the start button to begin your session. After you click start, you will see a timer in the upper right-hand corner.
- To navigate back to your list of sessions, click in the drop-down menu where you see your name and select “my reservations.”
- To log out, click the upper right-hand side menu and select “Log out.” You may log out while your session is in process. On the logout screen, you will see your list of active sessions.
To end your session:
- Login to the core kiosk using your iLab credentials.
- Find your current reservation in the list under “My kiosk sessions” and click the blue finish button.
- Confirm your action and click “finish session.” Your time on the instrument has been logged.
For questions, visit our FAQ page.
Recent Publications
Microglial microRNAs mediate sex-specific responses to tau pathology. A. Hsu, Q. Duan, S. McMahon, Y. Huang, S. A. Wood, N. S. Gray, B. Wang, B. G. Bruneau and S. M. Haldar. (2020). Salt-inducible kinase 1 maintains HDAC7 stability to promote pathologic cardiac remodeling. J Clin Invest, doi:10.1172/JCI133753 • L. Kodama, E. Guzman, J. I. Etchegaray, Y. Li, F. A. Sayed, L. Zhou, Y. Zhou, L. Zhan, D. Le, J. C. Udeochu, C. D. Clelland, Z. Cheng, G. Yu, Q. Li, K. S. Kosik and L. Gan (2020). Nat Neurosci. 23(2), 167–171. doi:10.1038/s41593-019-0560-7
Tau Reduction Prevents Key Features of Autism in Mouse Models. C. Tai, C. W. Chang, G. Q. Yu, I. Lopez, X. Yu, X. Wang, W. Guo and L. Mucke (2020). Neuron. doi:10.1016/j.neuron.2020.01.038
Single-Cell Determination of Cardiac Microtissue Structure and Function Using Light Sheet Microscopy. D. Turaga, O. B. Matthys, T. A. Hookway, D. A. Joy, M. Calvert and T. C. McDevitt (2020). Tissue Eng Part C Methods. doi:10.1089/ten.TEC.2020.0020
Self-Organized Pluripotent Stem Cell Patterning by Automated Design. D. Briers, A.R.G. Libby, I. Haghighi, D.A. Joy, B.R. Conklin, C. Belta, T.C. McDevitt (2019). Cell. doi:10.2139/ssrn.3318933
A Mac2-positive progenitor-like microglial population survives independent of CSF1R signaling in adult mouse brain. L.Zhan, P.D. Sohn, Y. Zhou, Y. Li, L. Gan (2019). bioRxiv. doi:10.1101/722090.
Context-Specific Transcription Factor Functions Regulate Epigenomic and Transcriptional Dynamics during Cardiac Reprogramming. N.R. Stone, C.A. Gifford, R. Thomas, K.J.B. Pratt, K. Samse-Knapp, T.M.A. Mohamed, E.M. Radzinsky, A. Schricker, L. Ye, P. Yu, J.G. van Bemmel, K.N. Ivey, K.S.Pollard, D. Srivastava (2019). Cell Stem Cell. 25(1), 87–102.e9
Single-cell analysis of cardiogenesis reveals basis for organ-level developmental defects. T.Y. de Soysa, S.S. Ranade, S. Okawa, S. Ravichandran, Y. Huang, H.T. Salunga, A. Schricker, A. del Sol, C.A. Gifford, D. Srivastava (2019). Nature. 572, 120–124.
Premature MicroRNA-1 Expression Causes Hypoplasia of the Cardiac Ventricular Conduction System. E. Samal, M. Evangelista, G, Galang, D. Srivastava, Y. Zhao, V. Vedantham (2019). Frontiers in Physiology. doi: 10.3389/fphys.2019.00235
Gladstone-led Publications
Fibrinogen in neurological diseases: mechanisms, imaging and therapeutics. M. A. Petersen, J. K. Ryu and K. Akassoglou (2018). Nat Rev Neurosci. 19(5), 283–301. doi:10.1038/nrn.2018.13
Microglial microRNAs mediate sex-specific responses to tau pathology. L. Kodama, E. Guzman, J. I. Etchegaray, Y. Li, F. A. Sayed, L. Zhou, Y. Zhou, L. Zhan, D. Le, J. C. Udeochu, C. D. Clelland, Z. Cheng, G. Yu, Q. Li, K. S. Kosik and L. Gan (2020). Nat Neurosci. 23(2), 167–171. doi:10.1038/s41593-019-0560-7
Do Microglial Sex Differences Contribute to Sex Differences in Neurodegenerative Diseases? L. Kodama and L. Gan (2019). Trends Mol Med. 25(9), 741–749. doi:10.1016/j.molmed.2019.05.001
Differential effects of partial and complete loss of TREM2 on microglial injury response and tauopathy. F. A. Sayed, M. Telpoukhovskaia, L. Kodama, Y. Li, Y. Zhou, D. Le, A. Hauduc, C. Ludwig, F. Gao, C. Clelland, L. Zhan, Y. A. Cooper, D. Davalos, K. Akassoglou, G. Coppola and L. Gan (2018). Proc Natl Acad Sci U S A. 115(40), 10172–10177. doi:10.1073/pnas.1811411115
Proximal recolonization by self-renewing microglia re-establishes microglial homeostasis in the adult mouse brain. L. Zhan, G. Krabbe, F. Du, I. Jones, M. C. Reichert, M. Telpoukhovskaia, L. Kodama, C. Wang, S. H. Cho, F. Sayed, Y. Li, D. Le, Y. Zhou, Y. Shen, B. West and L. Gan (2019). PLoS Biol. 17(2), e3000134. doi:10.1371/journal.pbio.3000134
Direct Reprogramming of Human Fibroblasts toward a Cardiomyocyte-like State. J.D. Fu, N.R. Stone, L. Liu, C.I Spencer, L. Qian, Y. Hayashi, P. Delgado-Olguin, S. Ding, B.G. Bruneau, D. Srivastava (2013). Stem Cell Reports. 1, 235–247
Context-Specific Transcription Factor Functions Regulate Epigenomic and Transcriptional Dynamics during Cardiac Reprogramming. N. R. Stone, C. A. Gifford, R. Thomas, K. J. B. Pratt, K. Samse-Knapp, T. M. A. Mohamed, E. M. Radzinsky, A. Schricker, L. Ye, P. Yu, J. G. van Bemmel, K. N. Ivey, K. S. Pollard and D. Srivastava (2019). Cell Stem Cell. 25(1), 87–102 e109. doi:10.1016/j.stem.2019.06.012
Chemical Enhancement of In Vitro and In Vivo Direct Cardiac Reprogramming. T. M. Mohamed, N. R. Stone, E. C. Berry, E. Radzinsky, Y. Huang, K. Pratt, Y. S. Ang, P. Yu, H. Wang, S. Tang, S. Magnitsky, S. Ding, K. N. Ivey and D. Srivastava (2017). Circulation. 135(10), 978–995. doi:10.1161/CIRCULATIONAHA.116.024692
Fibrinogen induces neural stem cell differentiation into astrocytes in the subventricular zone via BMP signaling. L. Pous, S. S. Deshpande, S. Nath, S. Mezey, S. C. Malik, S. Schildge, C. Bohrer, K. Topp, D. Pfeifer, F. Fernandez-Klett, S. Doostkam, D. K. Galanakis, V. Taylor, K. Akassoglou and C. Schachtrup (2020). Nat Commun. 11(1), 630. doi:10.1038/s41467-020-14466-y
Imaging the dynamic interactions between immune cells and the neurovascular interface in the spinal cord. N. Borjini, E. Paouri, R. Tognatta, K. Akassoglou and D. Davalos (2019). Exp Neurol. 322(113046). doi:10.1016/j.expneurol.2019.113046
Chromatin and epigenetics in development: a Special Issue. B. G. Bruneau, H. Koseki, S. Strome and M. E. Torres-Padilla (2019). Development. 146(19), doi:10.1242/dev.185025
Genome of the Komodo dragon reveals adaptations in the cardiovascular and chemosensory systems of monitor lizards. A. L. Lind, Y. Y. Y. Lai, Y. Mostovoy, A. K. Holloway, A. Iannucci, A. C. Y. Mak, M. Fondi, V. Orlandini, W. L. Eckalbar, M. Milan, M. Rovatsos, I. G. Kichigin, A. I. Makunin, M. Johnson Pokorna, M. Altmanova, V. A. Trifonov, E. Schijlen, L. Kratochvil, R. Fani, P. Velensky, I. Rehak, T. Patarnello, T. S. Jessop, J. W. Hicks, O. A. Ryder, J. R. Mendelson, 3rd, C. Ciofi, P. Y. Kwok, K. S. Pollard and B. G. Bruneau (2019). Nat Ecol Evol. 3(8), 1241–1252. doi:10.1038/s41559-019-0945-8
CTCF confers local nucleosome resiliency after DNA replication and during mitosis. N. Owens, T. Papadopoulou, N. Festuccia, A. Tachtsidi, I. Gonzalez, A. Dubois, S. Vandormael-Pournin, E. P. Nora, B. G. Bruneau, M. Cohen-Tannoudji and P. Navarro (2019). Elife. 8, doi:10.7554/eLife.47898
RNA Interactions Are Essential for CTCF-Mediated Genome Organization. R. Saldana-Meyer, J. Rodriguez-Hernaez, T. Escobar, M. Nishana, K. Jacome-Lopez, E. P. Nora, B. G. Bruneau, A. Tsirigos, M. Furlan-Magaril, J. Skok and D. Reinberg (2019). Mol Cell. 76(3), 412–422 e415. doi:10.1016/j.molcel.2019.08.015
A De Novo Shape Motif Discovery Algorithm Reveals Preferences of Transcription Factors for DNA Shape Beyond Sequence Motifs. M. A. H. Samee, B. G. Bruneau and K. S. Pollard (2019). Cell Syst. 8(1), 27–42 e26. doi:10.1016/j.cels.2018.12.001
Fibrinogen Activates BMP Signaling in Oligodendrocyte Progenitor Cells and Inhibits Remyelination after Vascular Damage. M. A. Petersen, J. K. Ryu, K. J. Chang, A. Etxeberria, S. Bardehle, A. S. Mendiola, W. Kamau-Devers, S. P. J. Fancy, A. Thor, E. A. Bushong, B. Baeza-Raja, C. A. Syme, M. D. Wu, P. E. Rios Coronado, A. Meyer-Franke, S. Yahn, L. Pous, J. K. Lee, C. Schachtrup, H. Lassmann, E. J. Huang, M. H. Han, M. Absinta, D. S. Reich, M. H. Ellisman, D. H. Rowitch, J. R. Chan and K. Akassoglou (2017). Neuron. 96(5), 1003–1012 e1007. doi:10.1016/j.neuron.2017.10.008
Heart enhancers with deeply conserved regulatory activity are established early in zebrafish development. X. Yuan, M. Song, P. Devine, B. G. Bruneau, I. C. Scott and M. D. Wilson (2018). Nat Commun. 9(1), 4977. doi:10.1038/s41467-018-07451-z
Cooperative activation of cardiac transcription through myocardin bridging of paired MEF2 sites. C. M. Anderson, J. Hu, R. Thomas, T. B. Gainous, B. Celona, T. Sinha, D. E. Dickel, A. B. Heidt, S. M. Xu, B. G. Bruneau, K. S. Pollard, L. A. Pennacchio and B. L. Blac. (2017). Development. 144(7), 1235–1241. doi:10.1242/dev.138487
An open-source control system for in vivo fluorescence measurements from deep-brain structures. S. F. Owen and A. C. Kreitzer (2019). J Neurosci Methods. 311(170–177). doi:10.1016/j.jneumeth.2018.10.022
Thermal constraints on in vivo optogenetic manipulations. S. F. Owen, M. H. Liu and A. C. Kreitzer (2019). Nat Neurosci. 22(7), 1061–1065. doi:10.1038/s41593-019-0422-3
Regulation of Tau Homeostasis and Toxicity by Acetylation. T. Tracy, K. C. Claiborn and L. Gan (2019). Adv Exp Med Biol. 1184(47-55). doi:10.1007/978-981-32-9358-8_4
An Alzheimer’s-disease-protective APOE mutation. K. A. Zalocusky, M. R. Nelson and Y. Huang. (2019). Nat Med, 25(11), 1648-1649. doi:10.1038/s41591-019-0634-9 23.
Editorial overview: Neurobiology of disease (2018). C. Bagni and A. C. Kreitzer (2018). Curr Opin Neurobiol. 48(iv-vi). doi:10.1016/j.conb.2018.01.005
Converging pathways in neurodegeneration, from genetics to mechanisms. L. Gan, M. R. Cookson, L. Petrucelli and A. R. La Spada (2018). Nat Neurosci. 21(10), 1300–1309. doi:10.1038/s41593-018-0237-7
A Subpopulation of Striatal Neurons Mediates Levodopa-Induced Dyskinesia. A. E. Girasole, M. Y. Lum, D. Nathaniel, C. J. Bair-Marshall, C. J. Guenthner, L. Luo, A. C. Kreitzer and A. B. Nelson (2018). Neuron. 97(4), 787–795 e786. doi:10.1016/j.neuron.2018.01.017
Motor thalamus supports striatum-driven reinforcement. Lalive AL, Lien AD, Roseberry TK, Donahue CH, Kreitzer AC. Elife. 2018;7:e34032. Published 2018 Oct 8. doi:10.7554/eLife.34032
Spatiotemporal distribution of fibrinogen in marmoset and human inflammatory demyelination. N. J. Lee, S. K. Ha, P. Sati, M. Absinta, N. J. Luciano, J. A. Lefeuvre, M. K. Schindler, E. C. Leibovitch, J. K. Ryu, M. A. Petersen, A. C. Silva, S. Jacobson, K. Akassoglou and D. S. Reich (2018). Brain. 141(6), 1637–1649. doi:10.1093/brain/awy082
Robust identification of deletions in exome and genome sequence data based on clustering of Mendelian errors. K. B. Manheimer, N. Patel, F. Richter, J. Gorham, A. C. Tai, J. Homsy, M. T. Boskovski, M. Parfenov, E. Goldmuntz, W. K. Chung, M. Brueckner, M. Tristani-Firouzi, D. Srivastava, J. G. Seidman, C. E. Seidman, B. D. Gelb and A. J. Sharp (2018). Hum Mutat. 39(6), 870–881. doi:10.1002/humu.23419
Fast-Spiking Interneurons Supply Feedforward Control of Bursting, Calcium, and Plasticity for Efficient Learning. S. F. Owen, J. D. Berke and A. C. Kreitzer (2018). Cell. 172(4), 683–695 e615. doi:10.1016/j.cell.2018.01.005
Pericytes: The Brain's Very First Responders? V. A. Rafalski, M. Merlini and K. Akassoglou (2018). Neuron. 100(1), 11–13. doi:10.1016/j.neuron.2018.09.033
Fibrin-targeting immunotherapy protects against neuroinflammation and neurodegeneration. J. K. Ryu, V. A. Rafalski, A. Meyer-Franke, R. A. Adams, S. B. Poda, P. E. Rios Coronado, L. O. Pedersen, V. Menon, K. M. Baeten, S. L. Sikorski, C. Bedard, K. Hanspers, S. Bardehle, A. S. Mendiola, D. Davalos, M. R. Machado, J. P. Chan, I. Plastira, M. A. Petersen, S. J. Pfaff, K. K. Ang, K. K. Hallenbeck, C. Syme, H. Hakozaki, M. H. Ellisman, R. A. Swanson, S. S. Zamvil, M. R. Arkin, S. H. Zorn, A. R. Pico, L. Mucke, S. B. Freedman, J. B. Stavenhagen, R. B. Nelson and K. Akassoglou (2018). Nat Immunol. 19(11), 1212–1223. doi:10.1038/s41590-018-0232-x
Military-related risk factors for dementia. H. M. Snyder, R. O. Carare, S. T. DeKosky, M. J. de Leon, D. Dykxhoorn, L. Gan, R. Gardner, S. R. Hinds, 2nd, M. Jaffee, B. T. Lamb, S. Landau, G. Manley, A. McKee, D. Perl, J. A. Schneider, M. Weiner, C. Wellington, K. Yaffe, L. Bain, A. M. Pacifico and M. C. Carrillo (2018). Alzheimers Dement. 14(12), 1651–1662. doi:10.1016/j.jalz.2018.08.011
A Genetically Encoded Fluorescent Sensor Enables Rapid and Specific Detection of Dopamine in Flies, Fish, and Mice. F. Sun, J. Zeng, M. Jing, J. Zhou, J. Feng, S. F. Owen, Y. Luo, F. Li, H. Wang, T. Yamaguchi, Z. Yong, Y. Gao, W. Peng, L. Wang, S. Zhang, J. Du, D. Lin, M. Xu, A. C. Kreitzer, G. Cui and Y. Li (2018). Cell. 174(2), 481–496 e419. doi:10.1016/j.cell.2018.06.042
Tau-mediated synaptic and neuronal dysfunction in neurodegenerative disease. T. E. Tracy and L. Gan (2018). Curr Opin Neurobiol. 51(134–138). doi:10.1016/j.conb.2018.04.027
The Psychiatric Cell Map Initiative: A Convergent Systems Biological Approach to Illuminating Key Molecular Pathways in Neuropsychiatric Disorders. A. J. Willsey, M. T. Morris, S. Wang, H. R. Willsey, N. Sun, N. Teerikorpi, T. B. Baum, G. Cagney, K. J. Bender, T. A. Desai, D. Srivastava, G. W. Davis, J. Doudna, E. Chang, V. Sohal, D. H. Lowenstein, H. Li, D. Agard, M. J. Keiser, B. Shoichet, M. von Zastrow, L. Mucke, S. Finkbeiner, L. Gan, N. Sestan, M. E. Ward, R. Huttenhain, T. J. Nowakowski, H. J. Bellen, L. M. Frank, M. K. Khokha, R. P. Lifton, M. Kampmann, T. Ideker, M. W. State and N. J. Krogan (2018). Cell. 174(3), 505–520. doi:10.1016/j.cell.2018.06.016
Klotho controls the brain-immune system interface in the choroid plexus. L. Zhu, L. R. Stein, D. Kim, K. Ho, G. Q. Yu, L. Zhan, T. E. Larsson and L. Mucke (2018). Proc Natl Acad Sci U S A. 115(48), E11388-E11396. doi:10.1073/pnas.1808609115
In Vivo Imaging of CNS Injury and Disease. K. Akassoglou, M. Merlini, V. A. Rafalski, R. Real, L. Liang, Y. Jin, S. E. Dougherty, V. De Paola, D. J. Linden, T. Misgeld and B. Zheng (2017). J Neurosci. 37(45), 10808–10816. doi:10.1523/JNEUROSCI.1826-17.2017
Core-led Publications
Single-Cell Determination of Cardiac Microtissue Structure and Function Using Light Sheet Microscopy. D. Turaga, O. B. Matthys, T. A. Hookway, D. A. Joy, M. Calvert and T. C. McDevitt (2020). Tissue Eng Part C Methods. doi:10.1089/ten.TEC.2020.0020
Lightsheet fluorescence microscopy of branching human fetal kidney. D. Isaacson, J. Shen, D. McCreedy, M. Calvert, T. McDevitt, G. Cunha and L. Baskin (2018). Kidney Int. 93(2), 525. doi:10.1016/j.kint.2017.09.010
Single-Cell Determination of Cardiac Microtissue Structure and Function Using Light Sheet Microscopy. D. Turaga, O. B. Matthys, T. A. Hookway, D. A. Joy, M. Calvert and T. C. McDevitt (2020). Tissue Eng Part C Methods. doi:10.1089/ten.TEC.2020.0020
Imaging the developing human external and internal urogenital organs with light sheet fluorescence microscopy. D. Isaacson, D. McCreedy, M. Calvert, J. Shen, A. Sinclair, M. Cao, Y. Li, T. McDevitt, G. Cunha and L. Baskin (2020). Differentiation. 111(12–21). doi:10.1016/j.diff.2019.09.006
Three-dimensional imaging of the developing human fetal urogenital-genital tract: Indifferent stage to male and female differentiation. D. Isaacson, J. Shen, M. Overland, Y. Li, A. Sinclair, M. Cao, D. McCreedy, M. Calvert, T. McDevitt, G. R. Cunha and L. Baskin (2018). Differentiation. 103(14–23). doi:10.1016/j.diff.2018.09.003
Equipment
Learn how our experts can support your research.
Lightsheet/Single Plane Illumination Microscopy
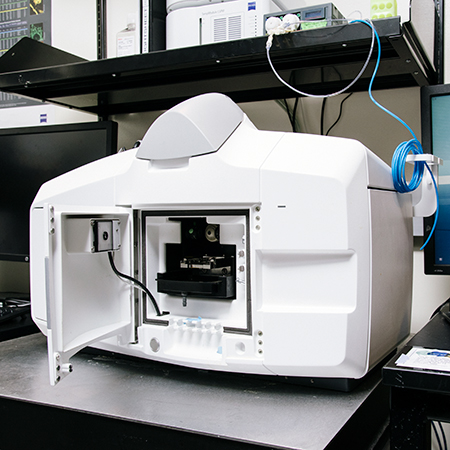

Fin Whale Zeiss Z1 Light Sheet Microscope
- Light Sheet Fluorescence Microscope
- For both aqueous and cleared samples
- Dual cameras for 2-color simultaneous acquisition (405nm, 488nm, 561nm, 640nm lasers)
- Sample requirements:
- Typically embedded in agarose; either aqueous or pre-cleared (Clarity, PACT, etc.)
- Samples usually fluorescently labeled
- Sample preparation can require extensive optimization
- Suitable applications:
- High-speed fixed or live-cell imaging (>100fps)
- Imaging large, thick samples at high resolution with extremely low-bleach, low phototoxicity
- Long-term live-cell imaging (>6 hours)
- Isotropic resolution- best possible 3D reconstruction
- Not good for:
- High magnification (max = 40x)
- Imaging large, thick samples at high resolution with extremely low-bleach, low phototoxicity
- Does the core provide training on this equipment? Yes
Miltenyi Blaze Ultra-Imaging 3D Light Sheet Microscope
Light Sheet Microscope:
- Immersion objectives: 1.1X and 4X (0.6X – 10X total range) Camera: 4.2 Mpx sCMOS (2048 x 2048 pixels)
- 5 lasers: 405, 488, 561, 639, 785nm
- Sample travel range : 24mm (X) x 50 mm (Y) x 23mm (Z)*
- Up to 6 light sheets (3 x 2 dual side) for optical sectioning
- Adjustable light sheet thickness: 4 – 24 μm (NA 0.0135 – 0.135)
*Z-range constrained by objective working distance (≥16 mm for 1.1X, ≥15 mm for 4X)
Sample types:
- Pre-cleared sample (aqueous buffers and organic solvents with RI 1.33 – 1.57)
- Organoids to adult mouse
Application:
- Imaging (multiple) small to large samples in 3D with low photobleaching and high relative speed
Not good for:
- Live imaging
- Structures needing to be resolved with greater that 10X magnification.
Confocal Microscopy
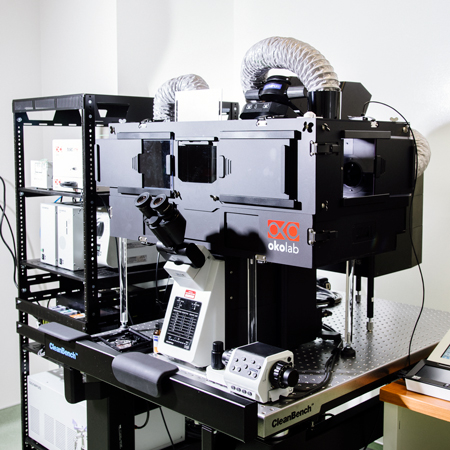

Snow Leopard (Olympus Fluoview FV3000)
- Specifications:
- Point-scanning confocal microscope
- Both Galvo & Resonance scanning for high speed imaging
- Multiple laser lines (405nm, 445nm, 488nm, 514nm, 594nm, 561nm, 638nm)
- Definite focus with Z drift correction function
- Inverted microscope with motorized (x,y,Z) stage
- >Incubation chamber (CO2, Temperature & Humidity) for both short & long live cell/tissue imaging
- Sample types
- Chamber slides, 35mm dishes or optical-quality well plates
- Combo 35mm & Chambered slide holder
- Both live & fixed tissues/cells
- Applications:
- Scanning confocal imaging for live and fixed -cell, organoid and tissue imaging
- Live-cell fluorescence dynamics (i.e. FRAP, FRET, GCaMP)
- Imaging individual cells at high resolution, any magnification
- Multipositional acquisition and tiling multiple images for large sample areas
- Not good for:
- Very thick samples (>100µm)
- Very high-speed imaging (>10fps)
Tarsier (Zeiss LSM880 Super-Resolution AiryScan Confocal Microscope)
- Specifications:
- Point-scanning confocal microscope
- Airyscan detector for increased resolution (>120nm lateral), sensitivity
- Multiple laser lines (405nm, 445nm, 488nm, 514nm, 561nm, 638nm)
- Definite focus
- Objective inverter for using dipping lenses
- Sample types:
- Chamber slides
- 35mm dishes
- Optical-quality well plates
- Applications:
- Scanning confocal imaging for mid-speed (13 fps) live-cell imaging
- Live-cell fluorescent dynamics (i.e. FRAP, FRET, GCaMP)
- Imaging individual cells at high resolution, any magnification
- Multipositional acquisition and tiling multiple images for large sample areas
- Not good for:
- Very thick samples (>100µm)
- Very high-speed imaging (>10fps)
Widefield Microscopy
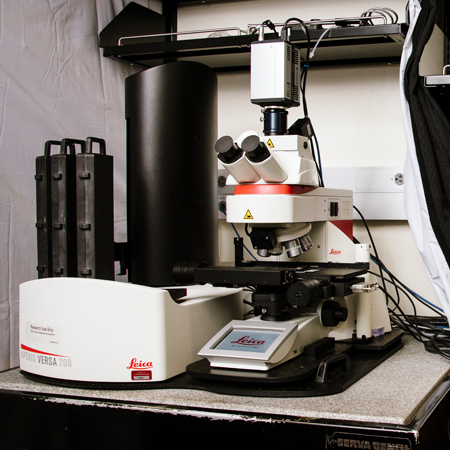






Octopus: Leica Versa 200 Automated Slide Scanner
- Specifications:
- Automated slide scanner with wide-field fluorescence and brightfield imaging
- Metal halide illumination; filters for DAPI, GFP, RFP, and Cy5
- High sensitivity RGB and monochrome camera
- 5x, 10x, 20x, 40x air objectives and 63x oil objective (with auto-oiler)
- Sample types:
- Slides / high throughput
- Applications:
- Automated whole slide imaging for up to 200 slides per run
- Multipositional acquisition and tiling for large sample areas
- Not good for:
- Long term live-cell imaging
- Thick samples (>50um)
- Does the core provide training on this equipment? Yes
Coyote 1 (Keyence epifluorescence BZX810 )
- Specifications:
- Automated inverted wide-field epifluorescence and brightfield imaging system
- Metal halide illumination; filters for DAPI, GFP, RFP, and Cy5
- High sensitivity RGB and monochrome camera
- Sample types:
- Slides or 35mm dishes
- Tissue culture plates
- Petri dishes
- Applications:
- Epifluorescence imaging for fixed and live-cell samples
- Multipositional acquisition and tiling multiple images for large sample areas
- Structured illumination for increased resolution (pseudo-confocal)
- Phase contrast and oblique illumination for high contrast brightfield imaging
- Not good for:
- Long term live-cell imaging
- Thick samples (>50um)
Coyote 2 & 3 (Keyence epifluorescence BZ9000)
- Specifications:
- Automated wide-field epifluorescence system
- Metal halide illumination; filters for DAPI, GFP, RFP, and Cy5
- Sample types:
- Slides
- 35mm dishes
- Applications:
- Epifluorescence imaging for fixed and live-cell samples
- Tiling multiple images for large sample areas
- Not good for:
- Long term live-cell imaging
- Thick samples (>50um)
- Plastic tissue culture plates
Cuttlefish (Zeiss AxioObserver Z1 Inverted Microscope)
- Specifications:
- Wide-field epifluorescence system for fixed and live-cell imaging
- Metal halide illumination; filters for DAPI, GFP, RFP, and Cy5
- Definite focus, motorized stage, and stable incubation (temperature and CO2 control)
- Polarizing filter for imaging birefringence
- Sample types:
- Slides
- 35mm dishes
- Tissue culture well plates
- Applications:
- Long-term live-cell imaging experiments
- Multipositional acquisition and tiling multiple images for large sample areas
- Not good for:
- High-speed live-cell imaging
- Thick samples (>50um)
Zeiss Axioscan 7 Slide scanner
- High End Brightfield imaging with Axiocam 705 color camera for high-resolution color acquisition
- High End Fluorescent imaging with Axiocam 712 monochrome camera for high-resolution monochrome acquisition
- High Performance Objectives: 5x, 10x/.45, 20x/0.8, 40x/.95
- DAPI, GFP, mcherry, Cy3, Cy5, Cy7
Sample type:
- Slides (brightfield or immunofluorescence)
Applications:
- Automated whole slide imaging
- 100 slide capacity magazine plus continuous Loading capability
Tissue Processor: AS


A82300001A Excelsior AS Tissue Processor with Line Conditioner
- Specifications:
- Automatically prepare tissue samples for laboratory testing, by fixing, dehydrating, clearing, and infiltrating them with paraffin
- Enhanced tissue quality
- Preheating reagents up to 35°C, minimizing tissue shock and facilitating reagent penetration
- Sample types:
- All types of tissues including fatty tissues
- Applications:
- Alcohol quality measurement enables you to extend your reagent life and provides significant cost savings
- Automatic reagent rotation further extends reagent life using a proven system
- Does the core provide training on this equipment? No
Epredia Histostar Embedding Station
Specifications:
- 5-liter paraffin capacity
- Cold plate area for 72 base molds
- Only workstation with heated wax trimmer built directly into the workspace
Does the core provide training on this equipment ? No
Microtomes

Leica RM2245 Automatic Microtome
- Specifications:
- The RM2245 is a semi-motorized rotary microtome, designed for routine in histopathology
- Manual sectioning is enhanced by a high-precision motorized specimen feed, which results in efficient operation with maximum section quality and reproducibility
- Application:
- Choose between conventional, full-hand-wheel rotation, manual sectioning or "rocking mode".
- Motorized sectioning and meets the many requirements of modern laboratories
- Does the core provide training on this equipment? Yes
Cryostats


CryoStar NX50 Cryostat Instrument (Epredia)
- Does the core provide training on this equipment? Yes
CryoStar NX70 Cryostat Instrument (Epredia)
- Specifications:
- A form-fitting design, integrated height control, and motorized sectioning
- Adjustable cutting speed from 0 mm/s to 256 mm/s
- Section thickness from 0.5 µm to 500 µm
- Vertical stroke length of 64 mm
- 48 mm horizontal feed range
- 20 µm specimen retraction on return stroke, with optical indication
- Applications:
- Mounting tissue to ready-to-cut with quick freeze and setup features
- Section tissue samples more efficiently with improved stability and individually controlled temperature settings on both the object holder and blade carrier
- Does the core provide training on this equipment? No
Coverslipper


Leica ST5020 Cover Slipper
- Specifications
- Integration into staining/coverslipping workstations to form a fully automated
- Coverslips at a rate of 300 slides per hour using standard microscope slides and glass coverslips of different sizes to increase productivity and efficiency in the lab.
Leica ST5020 Auto Stainer
Specifications:
- Automated deparaffinization, and H&E staining
- Reproducible and optimized protocol for staining
- Can stain up to 60 slides in one run
Does the core provide training on this equipment? No
Support Discovery Science
Your gift to Gladstone will allow our researchers to pursue high-quality science, focus on disease, and train the next generation of scientific thought leaders.
FAQs
How Do I Create an iLab Account?
If you're a UC San Francisco user, learn how to create an account with iLab.
If you’re part of a nonprofit organization or a for-profit company, you’ll need to sign a legal service agreement (LSA). Once the LSA is signed by both organizations, you’ll receive instructions on how to set up an iLab account to request services. Contact the core to start the LSA process.
How Can I Get a Quote for My Project?
- Login into your iLab account
- Initiate a service request
- Use the temporary PO number 11111 to finalize and submit your service request
- Email the core after your service request is submitted with the temporary PO to receive a formal quote
- Use the quote to generate a formal PO by your lab or organization’s financial department
- Use the valid PO to update your iLab account profile and billing or financial information
- Email the core a PDF copy of your formal PO
If you need help generating or submitting your PO, reach out to your lab’s grant or financial manager.
How Do I Get a Consultation?
How Do I Pay for Services?
When you create an iLab account, you will provide a valid PO. We do not accept credit cards or cash.
Who Can Access the Core’s Services and Instruments?
The core’s services are available to both Gladstone and outside researchers.
Can I Use the Core Equipment Independently?
Users can use some of the core equipment once they are trained and granted access by the core’s staff. Only formally trained users are allowed to use our core’s equipment.
What If I Already Have Experience Using Microscopes and Other Instruments?
In order to independently use the core’s microscopes and instruments, you must schedule a training session during which core staff will evaluate your experience and determine whether a second session is needed before gaining full access to the core’s equipment.
How Do I Request Services or Training?
Visit iLab and fill out the instrument training request. After you send the request, it typically takes 1–2 weeks to schedule a training session.
Our training protocol is a 2-step process. The first session is more general and covers care and handling of the microscope. The second session is focused on your specific project needs. At the end of these two training sessions, the core will evaluate your comfort level with the microscope before granting “assisted/dependent” or “unassisted/independent” user status with full access to the calendar.
How Do I Schedule or Make an Equipment Reservation?
How Do I Drop Off and Pick Up My Samples?
Sample drop-off
To drop off your samples, stop by the core located in room 367 at Gladstone Institutes, between 10am–3pm.
- FFPE samples:
- You should fix your samples (in 4 percent PFA or 10 percent formalin), rinse (3x) in 1x PBS, and store in 70 percent EtOH, before bringing to the core.
- Gradually increase EtOH ethanol concentration: 25 percent, 50 percent, and finally 70 percent EtOH approach is preferable or recommended.
- Frozen tissues: Samples can be brought to the core in one of the following conditions:
- In 30% sucrose (transported on ice)
- In OCT blocks or tissues slides (transported on dried ice)
Contact the core for the 10x Genomics Visium sample preparation
Sample pick-up
The core will let you know when your samples are ready. When they are ready for pick-up, stop by room 367 at Gladstone Institutes, between 10am–3pm.
Can Samples Be Sent by Mail?
All samples must be dropped off in person. To drop off your samples, stop by the core located in room 367 at Gladstone Institutes, between 10am–3pm
How Do I Know When My Samples Are Ready?
You will be notified by iLab mail. Pick up by the drop-off area, in room 367 at Gladstone Institutes. Remember to pick up your antibodies as well!
How Long Will It Take for My Request to Be Completed?
Standard turn-around time is typically 10–20 business days from the date the samples are dropped off and received by the core (not the project submission date via iLab), depending on the size of the order and how many requests we have in our queue.
Rush order turn-around is 5–10 days
Note that these are estimates and turn-around times can be longer due to the complexity and size of the project and our current workload.
Does the Core Provide Pathology Assessment?
Our core does not provide pathology assessment.
What Type of Tissue Can Be Submitted to the Core for Processing?
We accept a large variety of tissue types including regular soft tissues as well as bones, cells, and organoids pre-embedded in histogel or in suspension. All tissues must be fixed prior to submission.
Should I Fix My Samples Prior to Submitting Them to the Core, and What Is the Best Way to Fix Them?
Yes, all tissue should be fixed prior to submission. Most tissues do well with fresh perfusion followed by 4 percent PFA or 10 percent formalin fixation overnight in the fridge. Common exceptions are brain and bone samples, which require extended fixation.
Will Paraffin Blocks Be Returned after Completing the Request?
Yes.
When Should I Cite a Core Facility in a Publication, Presentation, or Poster?
We ask that you be generous with your citations. If you have any questions, reach out to one of the core managers.
Examples of when you should cite a core could include acknowledgement of:
- Space
- Equipment
- Purchased core reagents/media
- Services
- If a core staff member assists you in the design or execution of your experiments
Contact
Email@email for any information relating to POs, quotes, and administrative inquiries.
Email @email for any technical questions regarding projects.